HTLV-1 TUTORIAL
The HTLV-1 Molecular Epidemiology Database retrieves and stores annotated HTLV-1 proviral sequences from clinical, epidemiological, and geographical studies. It was developed through a partnership between Gonçalo Moniz Research Center/Oswaldo Cruz Foundation (Brazil), Bahia School of Medicine and Public Health (Brazil), Rega Institute at the Katholieke Universiteit Leuven (Belgium), and MyBioData (Belgium).
- Concerning the interface:
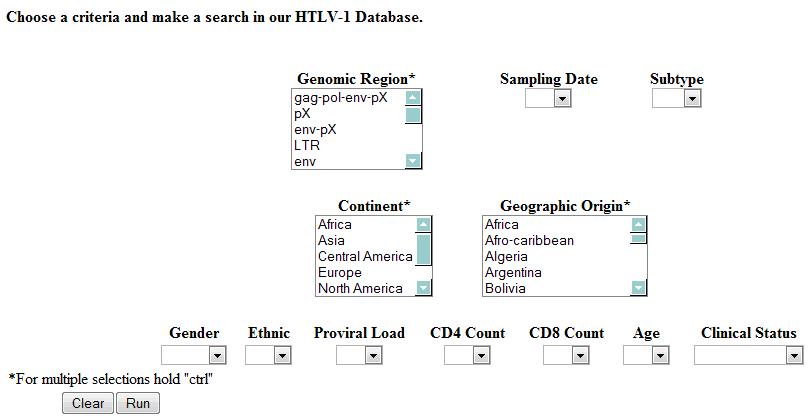
- The HTLV-1 Molecular Epidemiology Database homepage displays interface that contains numerous fields for refining database queries.
- One or more fields may be selected.
- The remaining unused fields will be ignored when searching the database, but their values will be presented in the final result.
- In the fields identified by the asterisk "*", the user may select multiple choices to make the search more efficient.
- Variable definitions:
- Genomic Region: shows all available genomic regions in the database.
- Sampling Date: values: YES (registered sampling date sequences), NO (unregistered sampling date sequences), and BLANK (If the user keeps the option selected, the search may return results with or without registered values to the referred variable).
- Subtype: shows all available subtypes in the database.
- Subgroup: this field will be visible only when the user selects Subtype "a" in the adjacent field. This variable shows all available subgroups in the database.
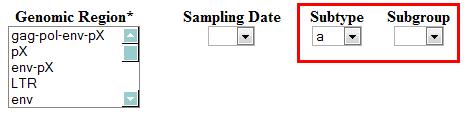
- Geographic Region: represents the geographic regions registered in the database.
- Continent: represents the registered continents in the database.
- Gender: values: MALE, FEMALE, and BLANK.
- Ethnic, Proviral load, CD4 and CD8 count, Age: values: YES (registered information sequences), NO (unregistered information sequences), and BLANK (If the user keeps the option selected, the search may return results with or without registered values to the referred variable).
- Clinical Status: Patients' clinic profile values when the sequence samples were collected (ASYMPTOMATIC, ATLL, TSH/HAM, OTHERS - other values than those which have been hereby described).
- Performing a Search:
Once the variables are set, the algorithm organizes the search as follows:
- Example 1: After a single value is set in different fields, the algorithm will search sequences in order of the first variable, the second variable, the third variable, and so forth.
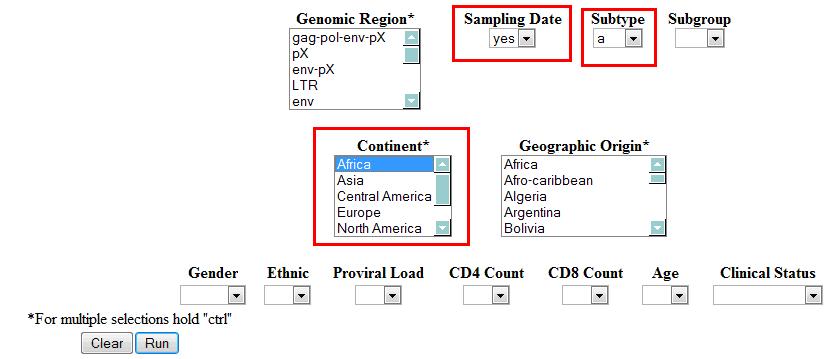 Example 1 will retrieve the (Subtype a) AND (Sampling Date) AND (Geographic Origin: Africa) sequence as a result
Example 1 will retrieve the (Subtype a) AND (Sampling Date) AND (Geographic Origin: Africa) sequence as a result
- Example 2: After two or more values from the same field are set, the algorithm will search sequences which contain (selection 1) OR (selection 2) OR (selection 3).
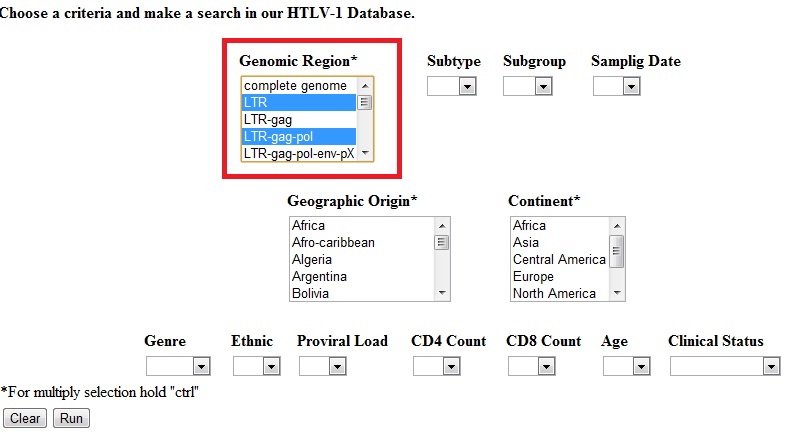 Example 2 will retrieve the (LTR sequences) OR (LTR-gag-pol sequences) as a result.
Example 2 will retrieve the (LTR sequences) OR (LTR-gag-pol sequences) as a result.
- Example 3: A combination of Example 1 and Example 2 will result in double "AND" selections.
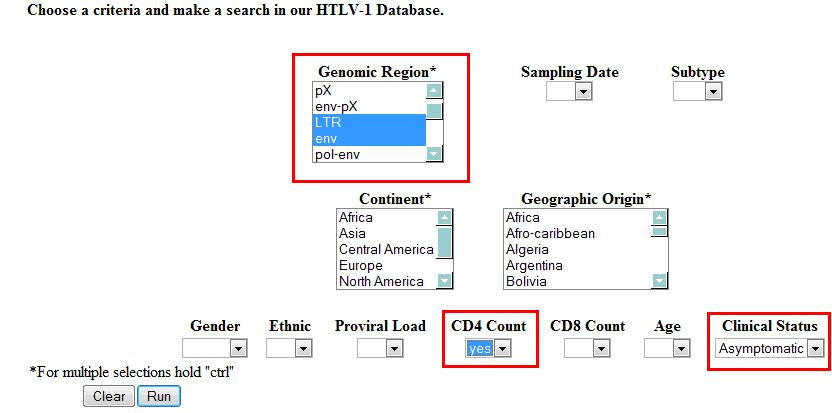 Example 3 will retrieve the (LTR or env) AND (CD4 Count) AND (Clinical Status: Asymptomatic)
Example 3 will retrieve the (LTR or env) AND (CD4 Count) AND (Clinical Status: Asymptomatic)
- Data Presentation:
After choosing the search criteria, click Run and the browser will be directed to a new page containing the results in table format:
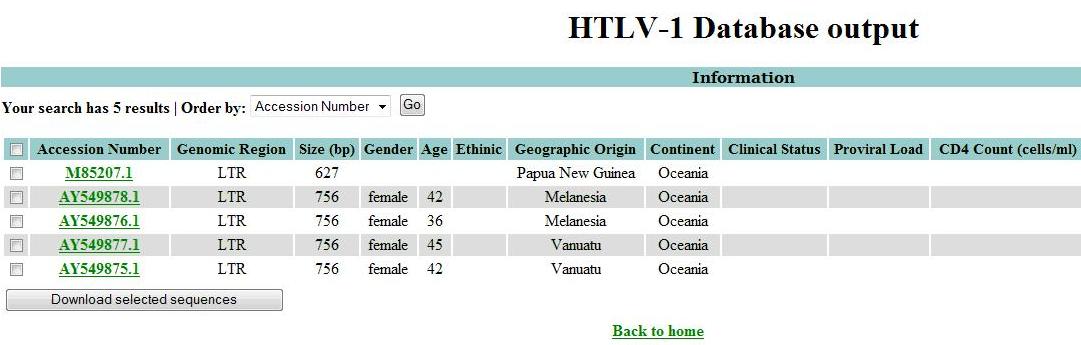
Next, select the sequences of interest. If the user want to select all sequences, he should click in the first checkbox.
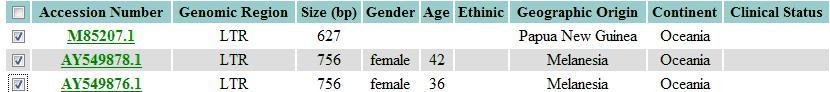
Next, the browser will be directed to the download page, where the sequence can be downloaded a FASTA or CSV file.

- Additional Comments
- The access numbers in the output page are actual links to the respective sequence in the GenBank page.
- The FASTA file features a header with the following structure:
>accession number.serial number.genomic region.isolated.base pair.subtype.subgroup (if available)
- The CSV file is a table containing all information presented in the search screen, along with a column called Sequence. That column matches the CSV file serial number, making it possible to relate the information in the CSV file with that in the FASTA file.
|





